Programmed bacteria target health and environmental challenges
Early detection of infectious diseases can be key to successful treatment, but we often don’t know we are infected until symptoms appear. What if our bodies could detect the presence of an infectious pathogen before the onset of disease?
Synthetic biology engineers are finding ways to do just that.
Synthetic biology is the design and construction of new biological parts and systems, and the re-design of existing and natural biological systems for specific purposes.
With an eye toward early disease detection, synthetic biology engineers at the University of Wisconsin have designed and engineered bacteria that find and detect fragments of DNA shed from infectious pathogens. Because the DNA of a pathogen in an individual could be identified before the appearance of symptoms, the system offers the promise of extremely early detection of deadly diseases such as sepsis where rapid detection is critical for successful treatment.
The work was done in the laboratory of Ophelia Venturelli, Ph.D., assistant professor of biochemistry, bacteriology, and chemical and biological engineering. The laboratory specializes in developing microbial control systems to address problems in medicine, agriculture, and the environment. A major area of investigation is how the human microbiome affects health and disease and how it can be modified to treat a variety of disorders. The work was co-led by postdoctoral fellow Yu-Yu Cheng and graduate student Zhengi Chen.
To design their bacterial DNA sensor, Venturelli’s team took advantage of the natural ability of a common bacterium Bacillus subtilis (B. subtilis) to capture DNA from its surroundings.
A series of genes was integrated into the B. Subtilis genome. This “genetic program” includes a region with a specific DNA sequence that matches the target organism. The researchers call this region a “cassette” because it is designed to be easily changed—popped in and out—with DNA sequences from the various organisms targeted for identification. The cassette region is part of a genetic program that is activated when B. subtilis finds the target sequence and brings it into the cell. Upon activation, the program directs a cascade of events that culminates in a fluorescent signal indicating that the bacterial sensor has detected the target organism.
In one of the first experiments, the team inserted a piece of DNA from the bacterium E. coli into the cassette region and tested the ability of the sensor bacteria to detect E. coli DNA mixed into the solution. When the E. coli DNA was brought into the cell it encountered its identical E. coli “cassette” DNA. Specialized bacterial proteins then swapped the two sequences, which activated the genetic program. The genetic program directs the production of green fluorescent protein (GFP) which created a strong fluorescent signal indicating that E. coli DNA was present in the sample.
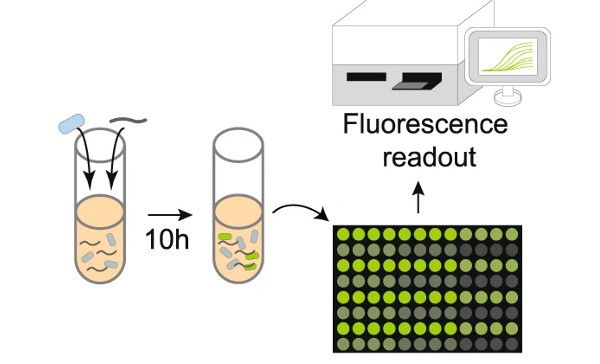
Having shown that their DNA sensor was able to detect the E. coli DNA, the Wisconsin team went on to test the ability of the system to identify human pathogens. Subsequent experiments demonstrated that the B. subtilis DNA sensor could be programmed to successfully detect a range of human bacterial pathogens. The human gut pathogen, Salmonella typhimurium (S. typhimurium), and the human skin and respiratory tract pathogen Staphylococcus aureus (S. aureus) were among those that were successfully detected by the B. subtilis DNA sensor.
“The ability of such sensors to detect human pathogens opens the possibility for a number of innovative diagnostic and therapeutic approaches,” explained Jermont Chen, Ph.D., Program Director at the National Institute of Biomedical Imaging and Bioengineering, which funded the study. “For example, the system could be used to rapidly diagnose and treat infectious diseases with specific, rather than broad spectrum antibiotics. This would be particularly valuable for high risk infections such as sepsis where time spent waiting for results of bacterial cultures rapidly decreases the likelihood of successful treatment.”
The laboratory is particularly interested in the microbiome—the millions of bacteria that reside in the human gut and throughout the body and interact with human cells in health and disease.
Venturelli elaborated on the potential use of the sensor in the human microbiome. “This work opens the possibility of designing sensors that harmlessly live in the ecosystem of the human body acting as sentinels for disease. The appearance of DNA from harmful bacteria growing in the gut, for example, could be detected by the B. subtilis sensor residing there, sending an early signal of a developing disease allowing for early treatment.”
Having successfully engineered the DNA sensor, the team is now focusing on improvements needed to move the technology towards practical application. These include increasing how efficiently B. subtilis takes-up DNA in its surrounding environment as well as increasing the efficiency of detecting pathogen DNA at low concentrations. Longer range goals include the development of more complex genetic programs allowing the sensing bacteria to identify a pathogen while also making the therapeutic proteins needed to immediately fight an infection.
The study was reported in the journal Nature Communications1. The work was supported by the National Institute Biomedical Imaging and Bioengineering grant R01EB030340 and the Defense Advanced Research Projects Agency (DARPA) grant HR0011-19-2-0002.
-Written by Thomas Johnson, Ph.D.
