Preclinical cancer treatment combines tumor-seeking bacteria with modified T cells
What if bacteria—which love to grow deep inside tumors—could guide cancer therapies directly to their target?
A team of researchers at Columbia University has engineered a bacterial strain to “light up” tumors so that reprogrammed T cells, drawn like a moth to a flame, can find and destroy them. Their preclinical treatment, reported in Science, could potentially be effective against any solid tumor type.
“While immunotherapy has shown enormous promise in treating a wide range of different cancers, it doesn’t work for all patients,” said Tuba Fehr, Ph.D., a program director in the Division of Discovery Science and Technology at NIBIB. “The treatment described here harnesses the tumor-seeking behavior of bacteria to selectively label cancer cells, allowing engineered T cells to effectively target a variety of solid tumors in mouse models. This tumor-agnostic approach could potentially overcome a major limitation of current CAR T-cell therapies.”
CAR T-cell treatment is a type of immunotherapy that modifies a patient's own immune cells to better identify and attack cancer cells within their body. Here’s how it works: T cells, taken from a patient, are engineered to express an artificial protein called a chimeric antigen receptor, or CAR. These artificial receptors are designed to recognize a specific antigen found on the surface of a tumor cell. Once infused back into the patient, the CAR T cells can find the tumor by binding to these antigens, ultimately leading to cancer cell death.
While CAR T-cell therapy is currently used to treat several different types of blood cancers, it hasn’t been approved to treat solid tumors. This is because a suitable solid tumor antigen for CAR T cells to recognize has yet to be identified. Tumors are heterogenous—meaning that the expression of different genes (including antigens) isn’t the same among all tumor cells. This can limit the efficacy of CAR T-cell therapy or lead to cancer relapse.
Instead of relying on an antigen that may or may not be expressed on the surface of a tumor cell, research in the Danino lab is taking a different approach. They are using an engineered bacterial strain, which selectively colonizes within the tumor core, to label the cancer cells with a synthetic antigen. These synthetic antigens act as a beacon, drawing CAR T cells that have been designed to recognize them to the tumor to effectively attack and destroy it.
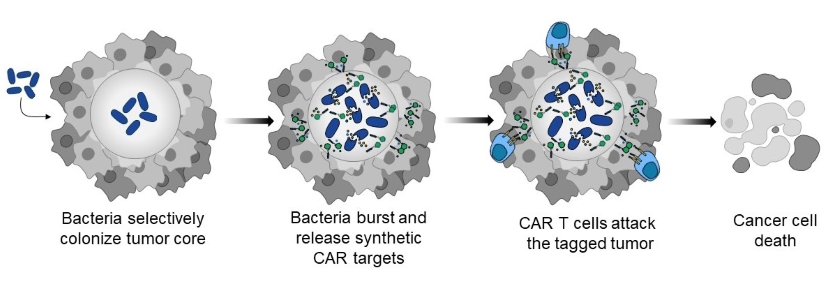
“Tumor cores are especially amenable to bacterial growth—things like low oxygen, low pH, necrotic tissue, and a lack of immune cells make up ideal conditions for bacterial colonization,” explained first study author Rosa Vincent, Ph.D. Taking advantage of these characteristics, the study authors engineered a non-pathogenic bacterial strain to release a fluorescent tag that their CAR T cells could recognize. “After the bacteria grow to a critical population density, they all simultaneously burst open and release their payload, which tags the tumor tissue and directs the CAR T cells towards them,” Vincent explained.
The researchers delivered their engineered bacteria intratumorally (by directly injecting the bacteria into tumors) or systemically (by administering the bacteria into the bloodstream) in several different mouse models. Both methods led to efficient tumor colonization, and, importantly, neither method led to bacterial growth in healthy organs. “During the course of our study, we didn’t see bacteria growing outside of the tumors, even in mice with severely compromised immune systems,” noted Vincent.
After the bacteria released their fluorescent antigen (roughly 48-72 hours after delivery), the researchers administered their engineered CAR T cells, either intratumorally or systemically. Following two rounds of CAR T-cell delivery, they found that their combinatorial treatment significantly reduced tumor growth in multiple mouse models, including models of breast and colorectal cancer.
“In addition to their ability to selectively target tumors, bacteria are immunogenic and can initiate a robust immune response, which likely enhances our treatment’s efficacy,” explained Vincent. Another benefit of using bacteria is their inability to easily grow outside of the tumor, she added.
“Identifying targetable tumor-specific antigens has been a significant obstacle in CAR T-cell research, so our team decided to design a synthetic antigen and program a CAR that could recognize it,” explained the study’s senior author Tal Danino, Ph.D., associate professor of biomedical engineering at Columbia University. “This bioengineering approach allowed us to circumvent a major bottleneck in the field.”
As this study was performed in mice, the study authors note that this experimental treatment is far from evaluation in clinical trials. Further, because humans are far more sensitive to bacteria than mice are, this combinatorial treatment will need to undergo rigorous safety studies before it is tested in human patients.
This study was supported by a grant from NIBIB (R01EB030352) along with a grant from the National Center for Advancing Translational Sciences (NCATS; UL1TR001873).
This science highlight describes a basic research finding. Basic research increases our understanding of human behavior and biology, which is foundational to advancing new and better ways to prevent, diagnose, and treat disease. Science is an unpredictable and incremental process—each research advance builds on past discoveries, often in unexpected ways. Most clinical advances would not be possible without the knowledge of fundamental basic research.
Study reference: Rosa L. Vincent et al. Probiotic-guided CAR-T cells for solid tumor targeting. Science 382, 211-218(2023). DOI:10.1126/science.add7034
