3D tissue samples, packed with data, can be analyzed by AI to predict outcomes
The analysis of human tissue samples—taking thin slices, staining them, mounting them on slides, and viewing them through a microscope—hasn’t fundamentally changed for more than a century. While this technique can help diagnose and predict disease progression, it is inherently limited by its two-dimensional nature. Humans, after all, aren’t flat, and 2D image slices can only tell us so much about our 3D bodies (and tissues contained therein).
3D pathology techniques have recently emerged, but their use in a clinical setting poses several challenges. Notably, the information captured from high-resolution 3D samples is enormously more complex than what is gathered from traditional 2D image slices. Analyzing 3D tissue datasets manually is nearly impossible and important details that could help improve prognosis are likely missed.
A team of NIBIB-funded researchers recently developed TriPath, an AI platform that can analyze 3D pathology images to predict disease outcomes. Their method, reported in the journal Cell, had improved performance in predicting prostate cancer outcomes when compared with traditional pathology approaches, such as analysis by expert pathologists using 2D images.
“Computational pathology is a booming field, and many algorithms are being developed to analyze tissue samples,” said study author Jonathan Liu, Ph.D., a professor in mechanical engineering at the University of Washington. “These success stories, based on 2D histology images, suggest that we could further revolutionize the field using 3D pathology datasets that are 100 times larger and that accurately capture the native morphology of the tissue.”
3D tissue sampling and AI analysis
Instead of thinly slicing and staining biopsied tissue, sample preparation for 3D images can be much simpler. Gentle approaches are available to render thick tissues transparent to light while maintaining the delicate structures and molecular constituents of the native tissue. Furthermore, larger tissue volumes may be imaged comprehensively, such as whole biopsies, and the sample may be stained with reversible dyes, or with no dye at all. In this study, the authors used two previously described methods—open-top light-sheet microscopy (OTLS) and micro-computed tomography (microCT)—to obtain 3D images from tissue biopsies and surgical specimens. While these imaging modalities are relatively expensive and are currently only used for research purposes, they can image tissues nondestructively and don’t require an experienced histotechnologist to prepare the samples, explained Liu.
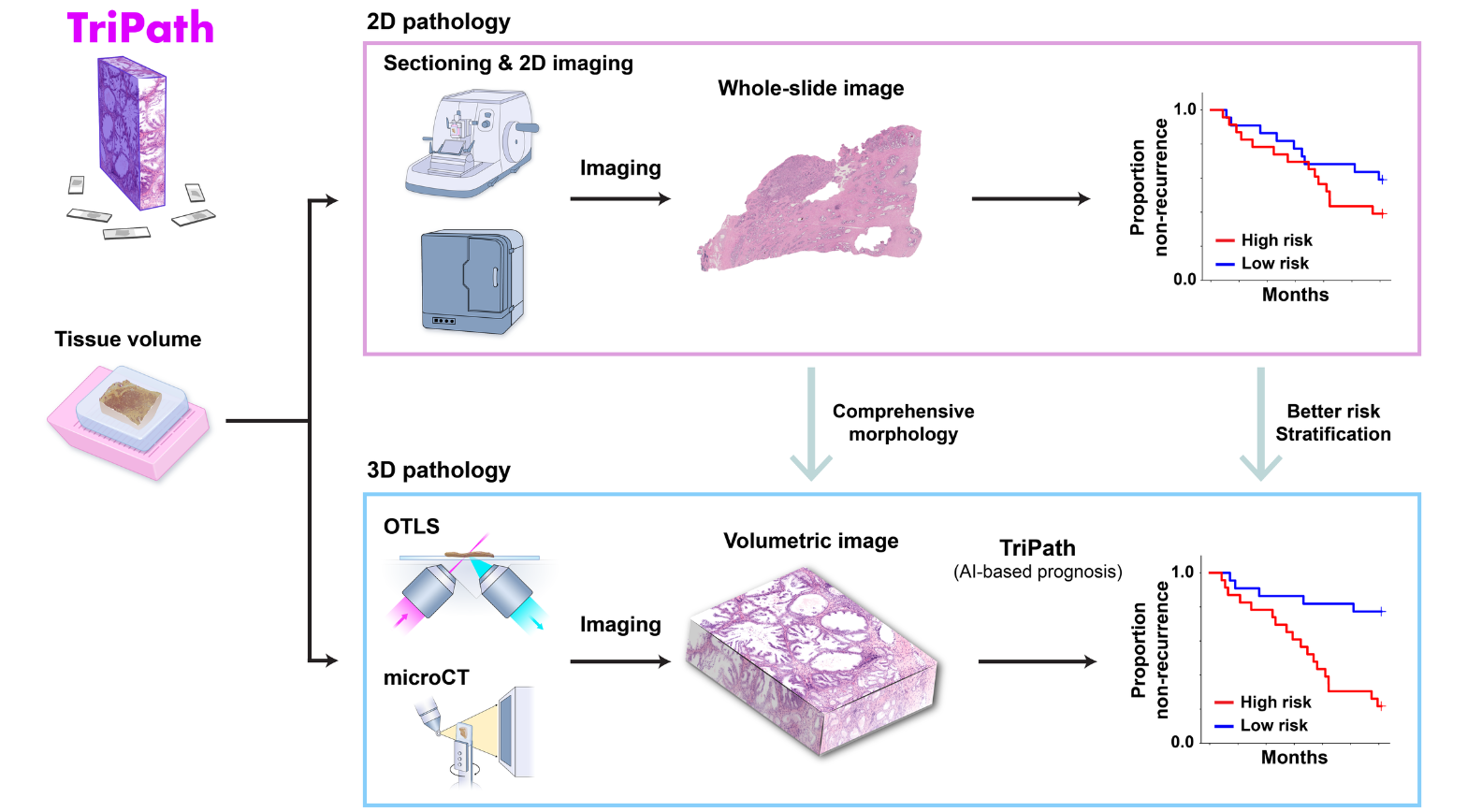
“Traditionally, pathologists look at several cross-sectional slices per biopsy, which may represent just 1% of the total biopsied tissue,” Liu said. “3D imaging techniques like OTLS and microCT can provide a more holistic view of the sample, allowing for the analysis of more tissue volume and also the discovery of microstructures and cell interactions that could improve clinical decision-making. Yet mining these immense datasets to extract insights for clinicians and patients has so far remained an unmet need.”
Liu and his colleagues at Harvard Medical School, including Andrew H. Song, Ph.D. and Faisal Mahmood, Ph.D., designed TriPath to predict clinical endpoints from 3D pathology images. This deep learning-based pipeline automatically divides the large, data-rich tissue into smaller segments, which are then analyzed individually. Specific predictive features within each segment are identified and weighted, and then information from each segment is combined together to provide an overall prognostic prediction.
Training for TriPath was “weakly supervised,” which is not to say that its predictions aren’t accurate. “Instead of using pixel-level annotations provided by skilled pathologists, we can train AI models based on ‘labels’ that apply to the whole specimen or the whole patient, such as survival or disease progression over time,” Liu explained. The ‘weak label’ in prostate cancer used to train TriPath was biochemical recurrence, or an increase in prostate-specific antigen (PSA) in the blood, within five years after patients with cancer had their prostates surgically removed. Biochemical recurrence is a potential indication that the cancer has returned.
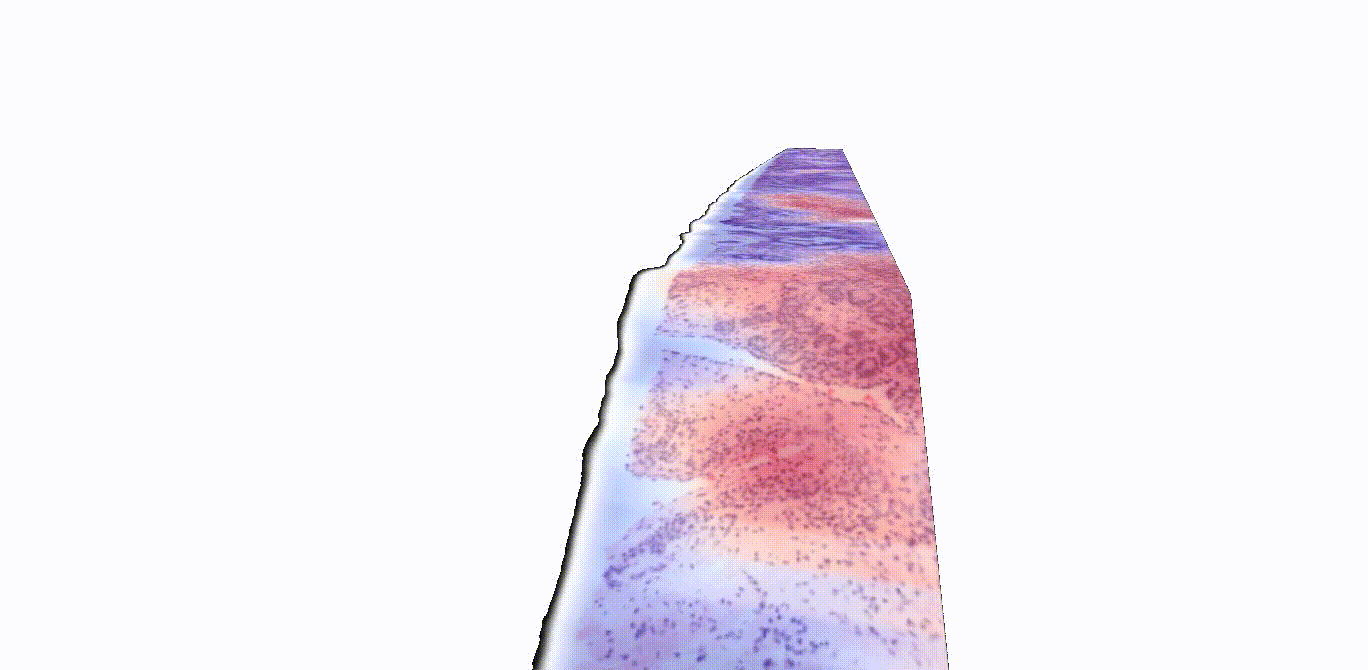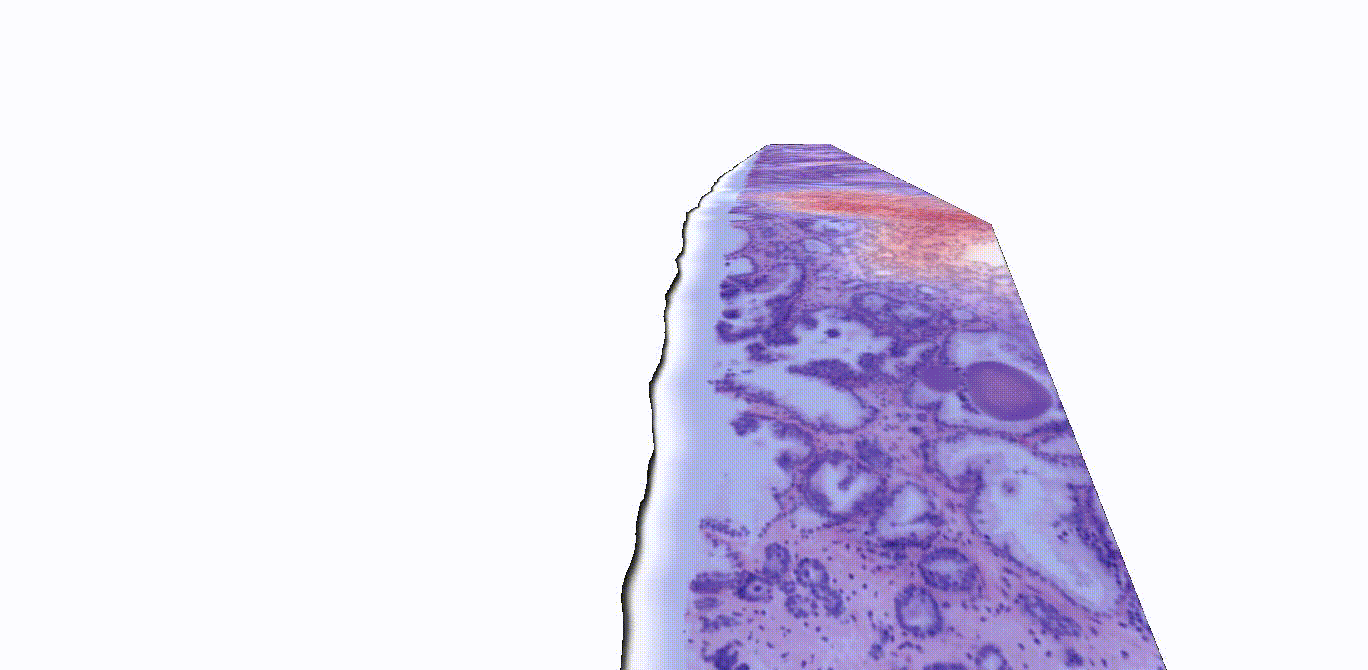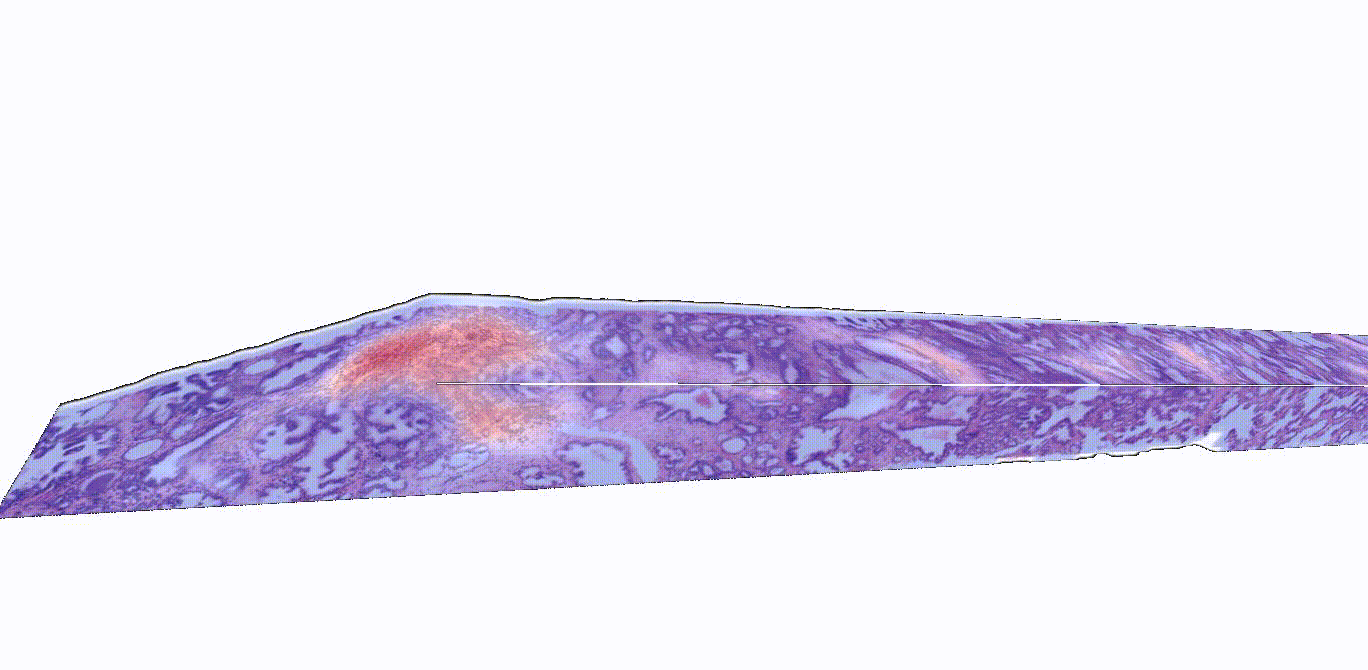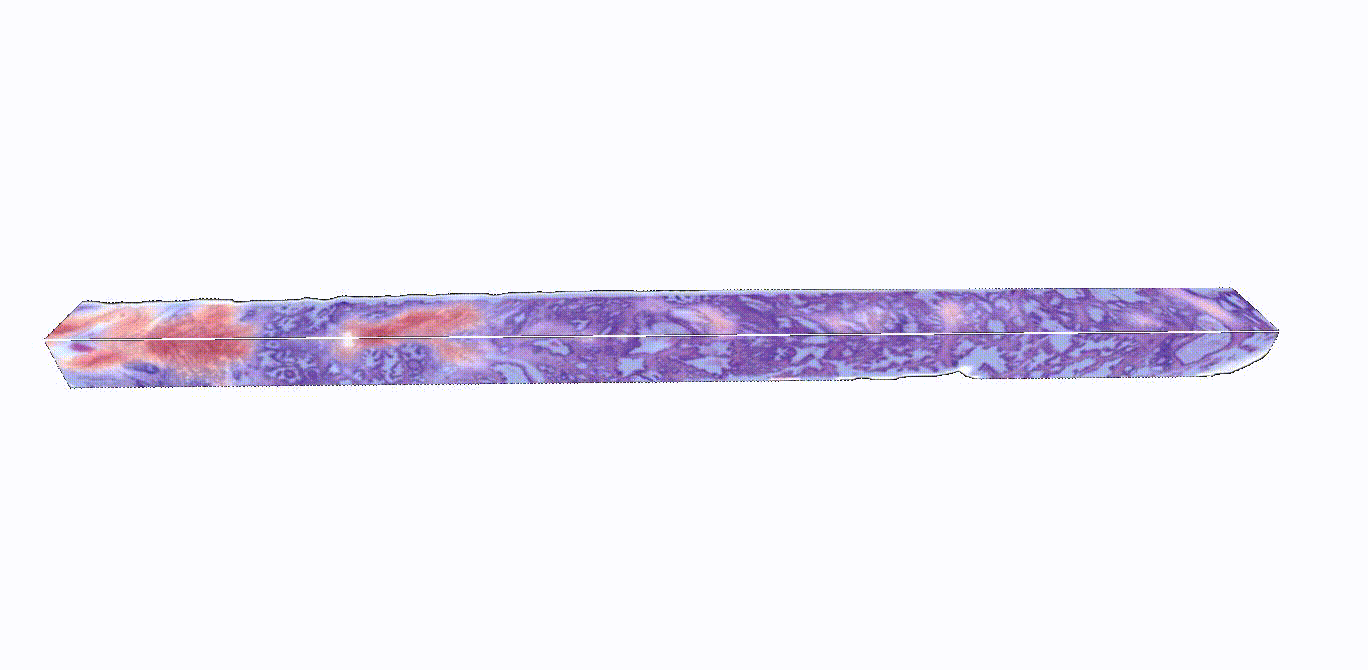
“A major challenge in prostate cancer care is risk stratification, as the majority of patients with this disease would never die from it,” said Liu. “If we can accurately identify the small fraction of patients with aggressive cancer that would most benefit from intensive therapies, we could spare the rest of the patients from unnecessary treatments that can have major side effects, like incontinence and impotence.”
Comparing TriPath to standard methods
TriPath was evaluated on its ability to predict biochemical recurrence using two separate groups of prostate cancer samples, one with images gathered via OTLS and the other with images gathered using microCT. The researchers found that TriPath could predict outcomes using either imaging modality, and that their models were the most accurate when TriPath analyzed more tissue volume and when the pipeline incorporated 3D morphological features from the tissue samples.
The researchers also compared the performance of TriPath to that of six expert genitourinary pathologists from four different countries. TriPath could more accurately predict biochemical recurrence than the expert pathologists—both when compared with each pathologist individually and when compared with their averaged predictions.
“Our results show that incorporating the native 3D morphology of the volumetric tissue sample can lead to improved prognostic predictions, which could ultimately improve patient care,” said Liu.
Looking ahead
While the analysis of 3D pathology samples is not currently part of routine clinical practice, Liu and team believe that this technology has the potential to augment 2D pathology approaches and improve outcomes. “This work provides initial evidence to support the value of 3D pathology and should spur additional and larger studies on computational 3D pathology in the future,” he said.
Future work will require the analysis of additional prostate tissue samples, collected from diverse locations and using different imaging techniques. The researchers also hope that this methodology can be applied to other types of cancer and diagnostic tasks. “We envision that TriPath will also evolve to reflect the rapid, breathtaking advances in machine learning and computer vision fields for even more improved prognostic performance,” said Song, who was the first author of this study.
“The computational pipeline described here has the potential to transform the way we analyze human tissue samples, unlocking invaluable information from everyday biopsies,” said Afrouz Anderson, Ph.D., a program director in the Division of Applied Science & Technology at NIBIB. “Further studies will be required to ensure that this technique performs accurately across multiple imaging modalities and all patient populations.”
This research was supported by grants from NIBIB (R01EB031002), the National Cancer Institute (NCI; R01CA268207 and T32CA251062), the National Institute of General Medical Sciences (NIGMS; R35GM138216), and the Office of the Director (S10OD023519). This study was also supported by a grant from the Department of Defense Prostate Cancer Research Program through W81WH-18-10358 and W81XWH-20-1-0851.
Study reference: Andrew H. Song et al. Analysis of 3D pathology samples using weakly supervised AI, Cell 187, 2502-2520, 2024. https://doi.org/10.1016/j.cell.2024.03.035
